Introducing Tahoe-x1

A 3-billion-parameter foundation model that learns unified representations of genes, cells, and drugs—achieving state-of-the-art performance across cancer-relevant single-cell biology benchmarks.
Scaling laws, meet cell biology
The last few years in AI have been defined by one simple pattern: scaling laws. More compute, more data, better models.
This has held true for language and protein modeling. But does it hold for systems biology—for models that learn how cells and genes function across contexts and perturbations like drugs?
Until now, two barriers have stood in the way:
- Lack of large, diverse single-cell data
- Lack of compute-efficient models that make billion-parameter exploration feasible
From Tahoe-100M to Tahoe-x1
With Tahoe-100M, we addressed the first barrier—building the largest perturbation dataset ever assembled: 100 million single cells across 50 cancer models and 1,100 drug perturbations.
It’s been downloaded nearly 200 K times in the past few months—a first for a biological dataset of this scale.
Now, we’re introducing Tahoe-x1 (Tx1) to tackle the second barrier.
What is Tahoe-x1?
Tx1 is the first billion-parameter, compute-efficient foundation model trained on perturbation-rich single-cell data. And it’s fully open-source, with open weights, training and evaluation code This makes it possible to empirically search for optimal architectures and hyperparameters for encooding cell states.
It’s also 3–30× more compute-efficient than previous cell-state models—pushing the frontier of what can be trained at this scale.

Engineering for scale and efficiency
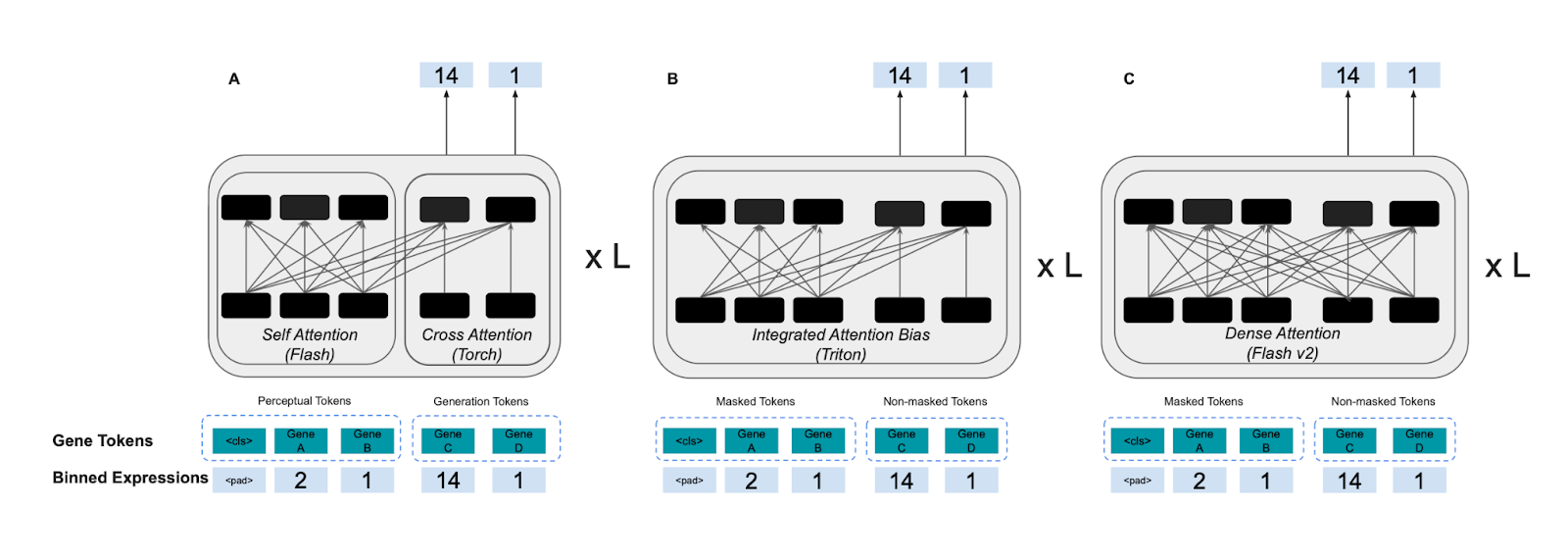
We borrowed the best tricks from large language models—FlashAttention v2, Fully Sharded Data Parallelism (FSDP), streaming datasets, and mixed precision training—and combined them with innovations tailored for biological modeling.
But even cooler: we redesigned the attention operation at the heart of these models.
In earlier Tx1 versions we built a Triton-optimized bias-matrix trick for masking, cutting GPU memory use ~10×. The final Tx1 design goes further: fully dense attention with FlashAttention v2—simpler, faster, and still highly memory-efficient.
A benchmark for discovery
We built Tx1 not only as a starting point for exploration, but also as a benchmark for the field. To measure what scaling really means for cell modeling, we designed new benchmarks focused on cancer discovery and translational tasks.
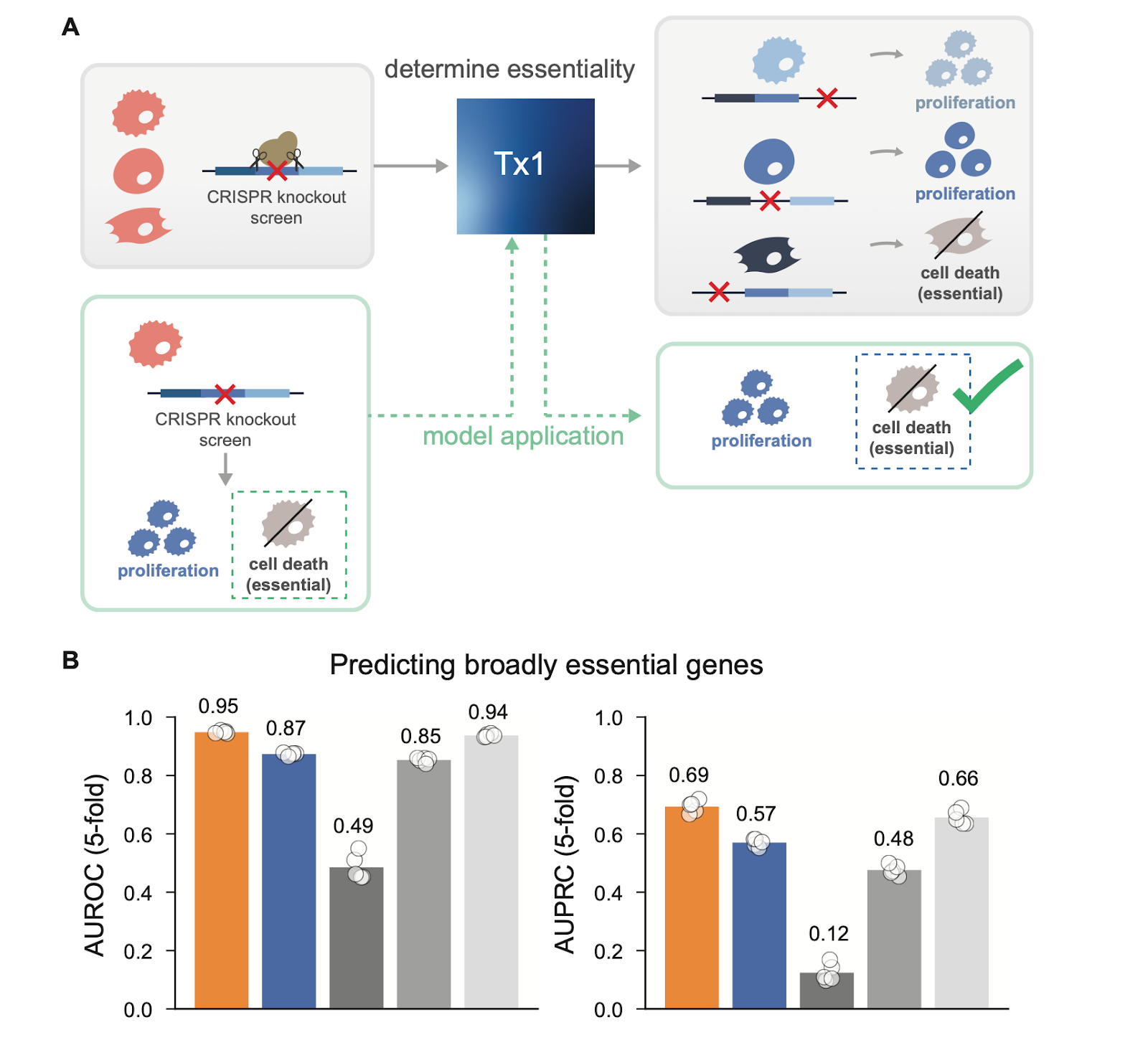
Predicting gene essentiality
Tx1 achieves state-of-the-art performance on predicting gene essentiality, as measured by the DepMap dataset—matching or surpassing linear baselines and outperforming all other models. This benchmark reflects the model’s ability to identify subtype-specific genetic dependencies, a crucial step toward new target discovery.
Inferring hallmark oncogenic programs
Similarly, Tx1 performs best on inferring hallmark oncogenic programs—a benchmark we built using MSigDB to evaluate how well a model captures the core transcriptional signatures of tumor progression.

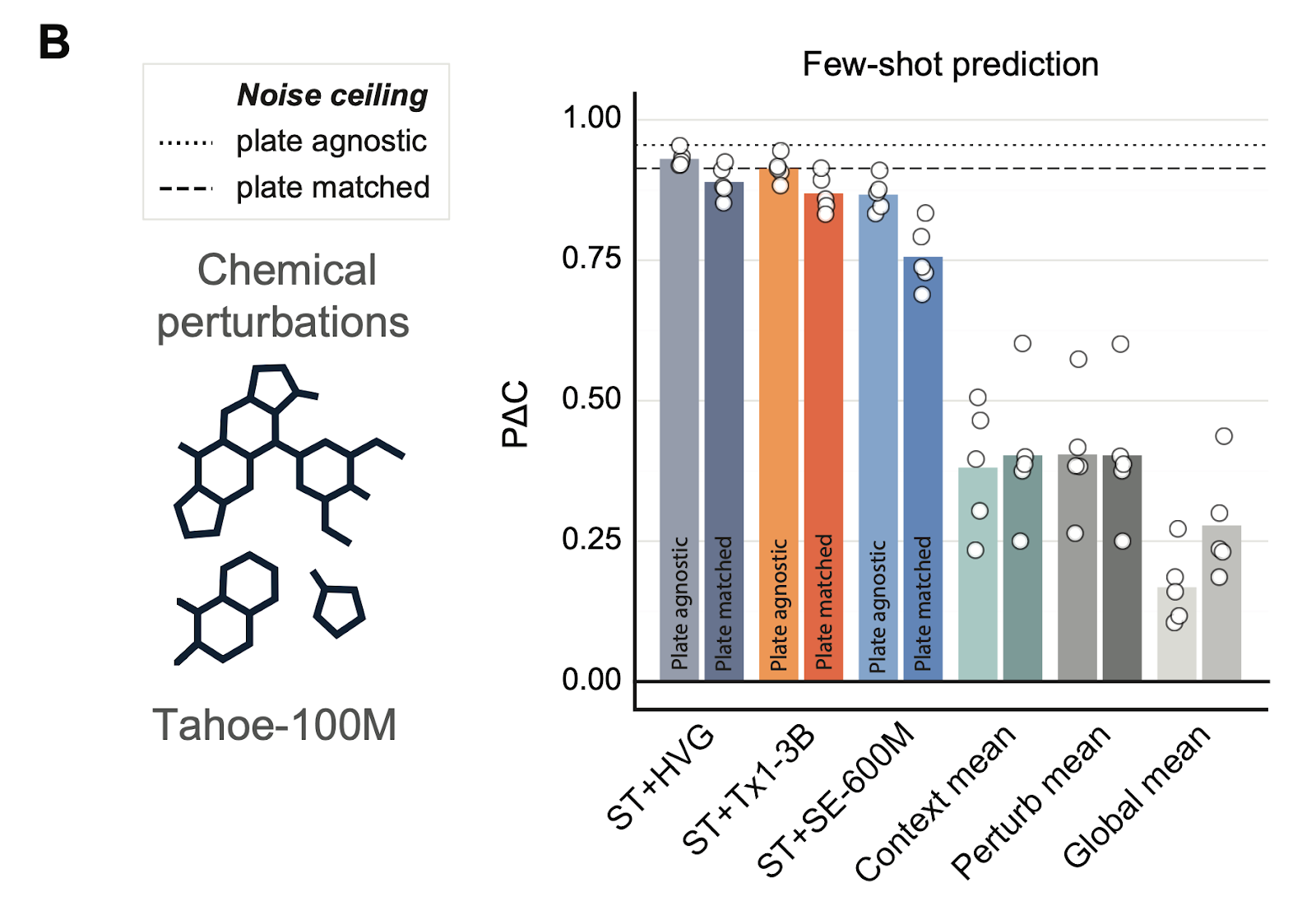
Toward in silico clinical trials
Tx1 also moves us closer to the goal of in silico clinical trials. Combined with post-training frameworks, it can predict drug responses in unseen cell types and patient contexts, generalizing across biological backgrounds.
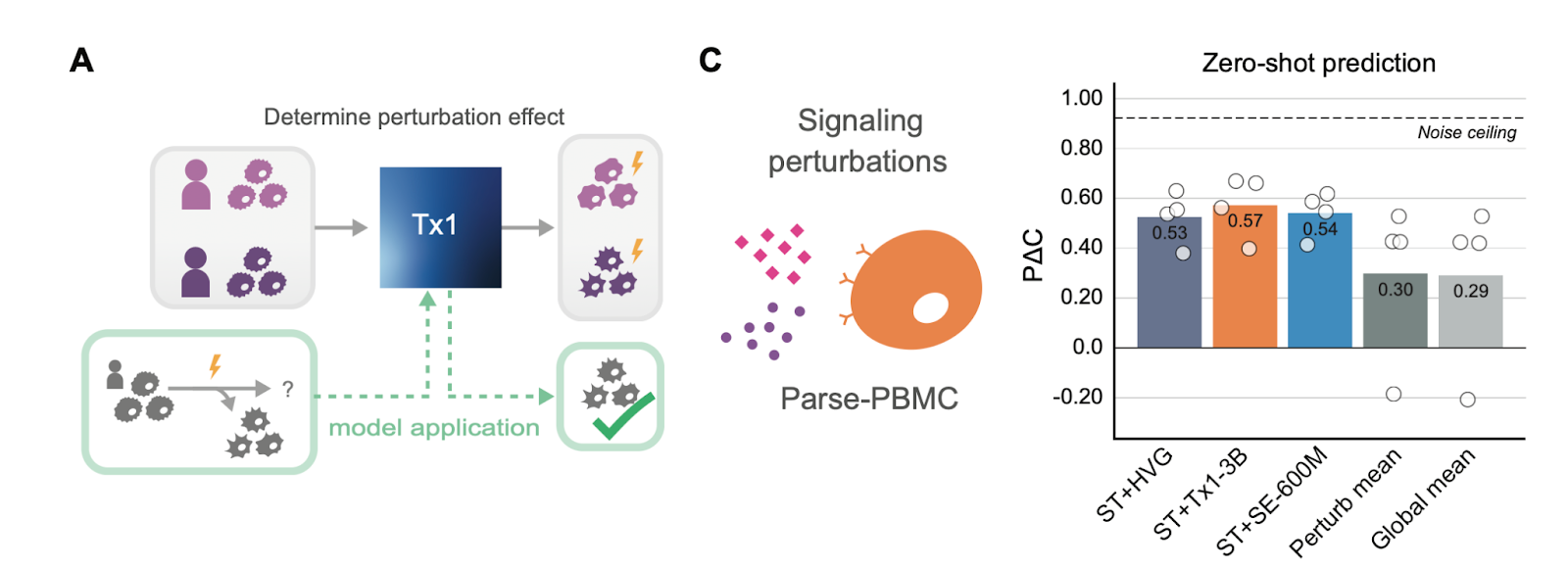
Zero-shot generalization, validated
Better embeddings mean better generalization. Tx1’s embeddings outperform other models in zero-shot prediction, confirming earlier findings from our friends at Arc Institute—that high-quality, perturbation-trained embeddings transfer robustly to new biological settings.
Try it yourself
We built a simple demo so you can upload your own single-cell transcriptomic data, generate Tx1 embeddings, and visualize them interactively.
See our DEMO on 🤗 Hugging Face and give it a try yourself!
Open source for collective progress
Progress won’t come from one model—it will come from hundreds of experiments, each testing new ways to represent the cell.
We built Tx1 for that purpose.
We’re open-sourcing everything—checkpoints, training code, and evaluation workflows—so others can build on top of it.
📄 See our preprint on biorxiv
🤗 See our Hugging Face model card
💻 See our GitHub repository